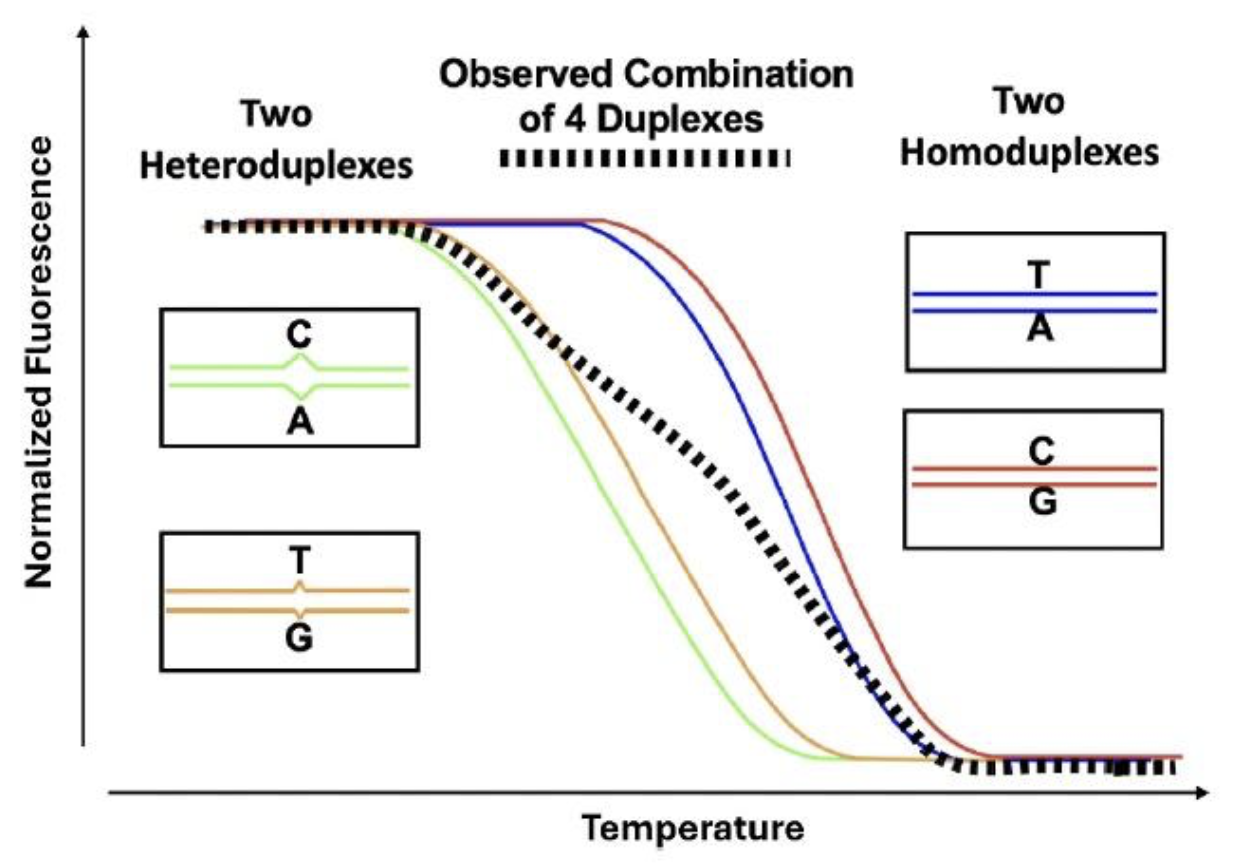
With high resolution melting and saturation dyes, it is possible to genotype without any probes. Using only 2 unlabeled primers, amplicon melting curves after PCR cluster into genotypes. Homozygotes are differentiated by melting temperature (Tm) and heterozygotes are identified by an altered curve shape caused by heteroduplexes (
Liew et al. 2004). After amplification of a heterozygote, 4 duplexes are formed, 2 homoduplexes and 2 heteroduplexes. The heteroduplexes are substantially lower in Tm, and the composite melting curve changes in shape (
Wittwer et al. 2003).
|
Idealized melting of a single base heterozygote after PCR amplification.
|

|
Predicted Tm difference between single nucleotide variant homozygotes.
|
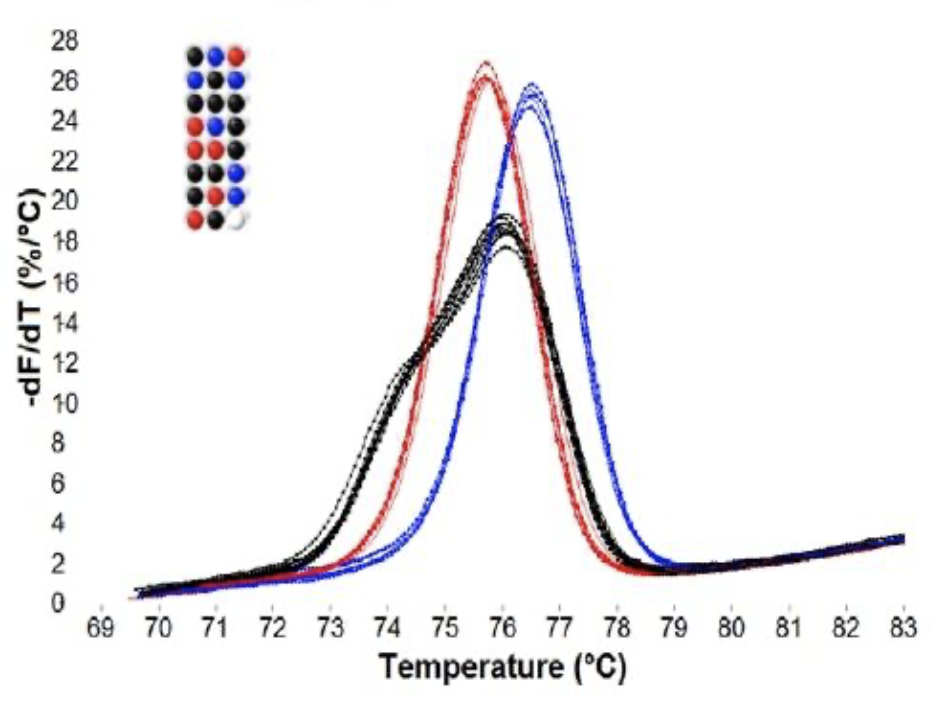
Heterozygotes are typically easy to identify by their shape, while homozygote differentiation depends on Tm shifts that may be small and require high resolution instruments and/or internal temperature controls (
Seipp et al. 2007). With small amplicons, most SNP homozygotes differ from WT homozygotes by about 1°C. Even SNP homozygotes that are predicted by nearest neighbor thermodynamic predictions to be identical to wild type can be distinguished (
Gundry et al. 2008). In many cases, different heterozygotes can be differentiated from each other (
Graham et al. 2005). When the PCR product is long enough, variants can be located to specific melting domains, as in the 544 bp product below.