| Top Software Resources
Fun Archives
|
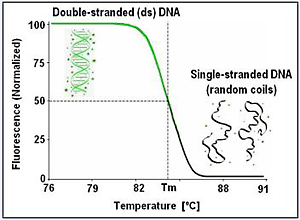
A Bit of History...In the 1960s, melting of double-stranded DNA was monitored by UV absorbance (hyperchromic effect). Analysis required µg amounts of DNA, and often took hours to complete, while samples were slowly heated at rates of 0.1-1.0 ºC/min. Contemporary DNA melting analysis uses fluorescence, and because it is a more sensitive method, it needs only ng amounts of DNA (amounts easily prepared by PCR). Fluorescent DNA melting analysis became popular with the 1997 advent of the real-time PCR instrument LightCycler®. Capillary sample formats and small sample volumes allowed better temperature control and fast melting rates of 0.1-1.0ºC/sec, shortening the melting time to a few minutes. The fluorescence indicator used back in 1997 was the dye SYBR® Green I, a sensitive, convenient dye for real-time monitoring of amplification and melting of amplified products.New to Melting? Watch the High-Resolution Melting Webinar with Dr. Carl Wittwer (Biotechniques, 2017) High-resolution melting analysis was introduced in 2003. Similar to the LightCycler, this too was a result of collaboration between our lab at the University of Utah and Idaho Technology, Inc (Salt Lake City, UT). The method requires high precision measurements of temperature and fluorescence to detect small sequence differences in PCR fragments, just by direct melting. Melting curve analysis for genotyping, mutation scanning, sequence matching, and copy number assessment were developed - analyses that traditionally required processing of PCR products by electrophoresis or other non-homogeneous means. Because of its speed and simplicity, the method rapidly grew in popularity. To read more, please follow the colored tabs in the navigation bar (top of page) .. Key references Contact us to receive reprints.
| |||||||||||||||||||||||
| © 2006- Wittwer Lab. All Rights Reserved. |