Melting analysis for relative copy number assessment can be performed with probes or dyes. Only PCR and melting of the targeted region and a reference sequence are required, although standards and interpolation are necessary.
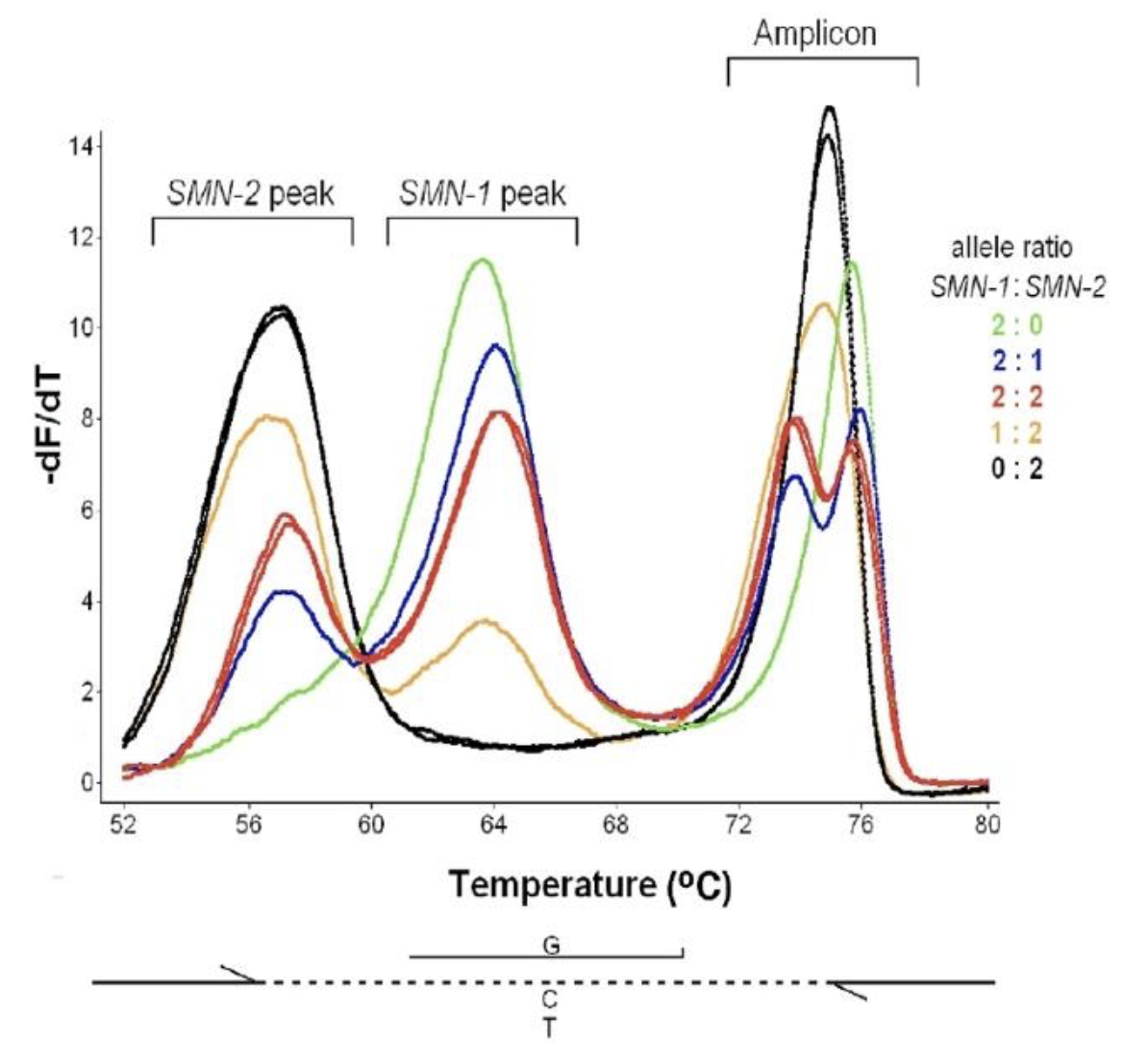
The figure on the left used unlabeled probes to assess copy number of SMN-1 and SMN-2 by asymmetric PCR using a common primer set and a saturating DNA dye in the presence of an unlabeled probe over a differentiating variant.
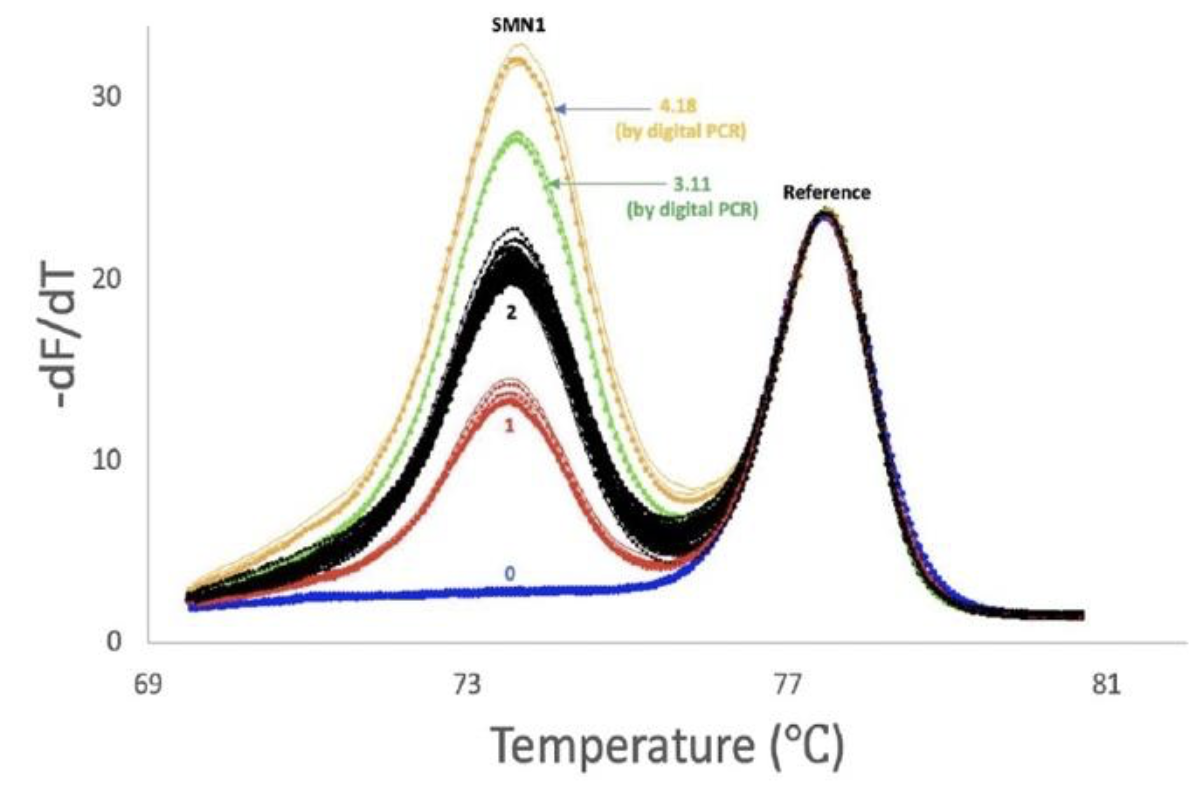
Relative copy numbers are apparent as peak height differences in both the low temperature probe region and the amplicon region. Another way to assess SMN-1 copy number is to amplify by allele-specific PCR along with a refence gene (figure on the right). To keep the relative proportions of target and reference constant during amplification, the dNTP concentration was limited (
Zhou et al. 2015).
The reference peaks are normalized both in temperature and fluorescence, resulting in clustering and quantification of the target peak. These methods are convenient, inexpensive, and rapid for targeted assessment of gene dosage for specific diagnoses, but they do not compete with copy number arrays or massively parallel sequencing when genome-wide enquiry is desired.
|
Copy number assessment of spinal muscular atrophy with unlabeled probes. An unlabeled probe targeted the C to T base change associated with the exon
7 deletion differentating SMN1 and SMN2. After asymmetric PCR, both unlabeled probe peaks and amplicon peaks correlate with SMN1:SMN2 ratios.
|
|
Copy number assessment of spinal muscular atrophy by PCR with restricted dNTPs. Duplex amplification of a reference gene (CFTR, exon 7) and
allele specific PCR of SMN1 was performed with 6.25 uM of each dNTP to equalize amplification of the 2 different targets. Copy numbers of 0,1,2,3 and 4 are clearly separated and confirmed by digital PCR.
|


