| Top Software Resources
Fun Archives
|
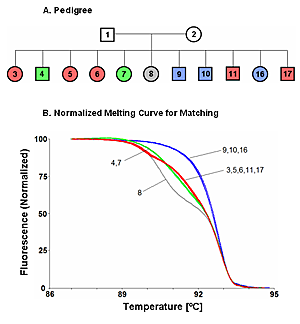
DNA matching - to quickly determine if two sequences are the sameIn some applications, knowing the exact complete genotype is less important than knowing whether the DNA sequences match. This occurs in tissue transplantation, genotype/phenotype correlations, and forensics.For example, in living-related organ transplants, siblings are usually tested for human leukocyte antigens (HLA) to obtain the best major histocompatibility match. This is done by serotyping, or genotyping HLA-A, B, C, DR (and often DQ) loci by laborious means. However, what is really important is to find a compatible sibling: that is, we need to know if there is a sibling with identical HLA sequence. For each available sibling, there is a 25% chance of a complete match. HLA serotyping or genotyping would not be needed if an HLA match can be confirmed by simpler means. By use of melting curves of PCR products, HLA sequence identity of siblings can quickly be established. Instead of using the melting temperature (or Tm) which is just one point in the melting curve, we use the information that is contained in the entire melting curve. The shape of the melting curve is used extensively in sequence matching as an indicator of heteroduplexes. A match in HLA sequence is suggested when two individuals have the same melting curve shapes. The match is confirmed by mixing the two DNAs at 1:1 ratio, remelting, and then comparing the new curve with the original curves from the individuals. If the sequences of the two samples are identical, the 1:1 mix is the same as the original melting curves. The figure shows an example of matching 11 siblings (CEPH family UT1331) using the HLA-A locus. The siblings separate into four matched groups. [Published in Zhou et al 2004] | ||||||||||||||||||||||||
| © 2006- Wittwer Lab. All Rights Reserved. |